Regents Professor and Head of Department
Microbiology and Molecular Genetics Cell: (405) 820-7329
Oklahoma State University Voice: (405) 744-6243
Stillwater, OK 74074 E-mail: tconway@okstate.edu
Professional Experience
College High School, Bartlesville, OK, 1975
B.S. 1979, Ph.D. 1984, Oklahoma State University (Microbiology)
Postdoctoral Research Associate, Oklahoma State University, 1985
Faculty Research Assistant (non-tenure track), University of Florida, 1985-1988
Assistant Professor of Biological Sciences, University of Nebraska, 1988 to 1993
Acting Associate Director, Center for Biotechnology, University of Nebraska, 1992-1993
Associate Professor of Biological Sciences, University of Nebraska, 1993-1994
Associate Professor of Microbiology, Ohio State University, 1994-1999
Associate Professor of Microbiology, University of Oklahoma, 1999-2004
Director, OU Microarray Core Facility, University of Oklahoma, 2000-2009
Director, OU Center for Informatics, University of Oklahoma, 2000-2009
Director, Oklahoma Bioinformatics Network (INBRE), statewide project, 2001-2009
Co-Director, OU Advanced Center for Genome Technology, 1999-2014
Professor of Microbiology, University of Oklahoma, 2004-2008
Henry Zarrow Presidential Professor, University of Oklahoma, 2008-2015
Professor of Microbiology, Oklahoma State University, 2015-2017
Head of Department of Microbiology, Oklahoma State University, 2015-
Regents Professor of Microbiology, Oklahoma State University, 2017-
Current Research Funding
RO1 GM117324 -01A1 3/1/17 to 1/31/23
NIH National Institute of General Medical Sciences $1,181,670
“Mechanisms of Nutrient Competition in the Intestine”
The major goal of this project is to determine mechanisms of nutrient competition between E.
coli strains in a mouse model of intestinal colonization.
3R01GM117324-01A1S1 10/1/17 to 10/31/20
NIH National Institute of General Medical Sciences $189,234
Diversity Supplement to parent project “Mechanisms of Nutrient Competition in the Intestine”
This award supports the postdoctoral training of Dr. Jerreme Jackson in accordance with the
NIGMS Diversity Supplement Program goal to promote diversity in the scientific research
workforce.
Previous Research Funding
1R01GM095370-01 01/01/11 to 12/31/14
NIH NIGMS $1,650,000 direct cost
“Symbiosis of E. coli and the Intestinal Microbiota in a Mouse Model”
RC1GM09207 09/30/09 to 8/31/12
NIH NIGMS $1,000,000 direct cost
“Comprehensive Mapping and Annotation of the E. coli Transcriptome”
2RO1-AI48945-05 6/01/04 to 5/31/09
National Institutes of Health $2,158,563 total cost
“Growth and Colonization of the Intestine by E. coli”
More than 25 grant awards from DOE, USDA, NREL, NSF, Genencor, and NIH
Continuously funded since 1990, totaling more than $17M
Current Research
We work on two main projects: to elucidate the mechanisms of nutrient acquisition by Escherichia coli in the intestine and to use single-nucleotide-resolved transcriptome data for comparative gene expression analysis of competing intestinal bacteria.
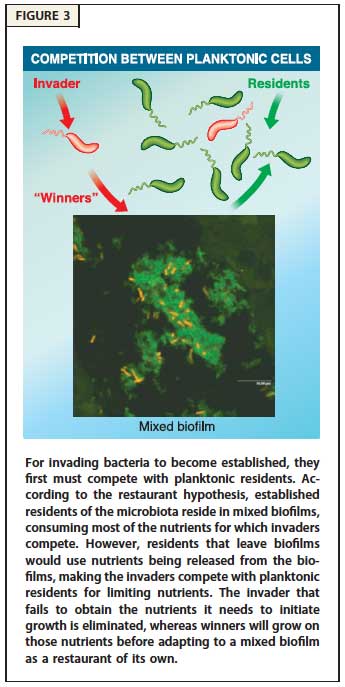
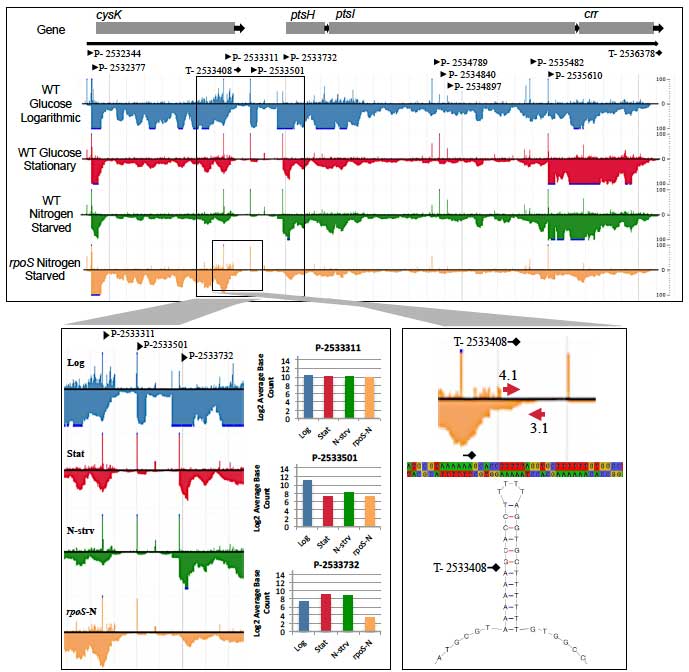
Mechanisms of Nutrient Competition in the Mammalian Intestine. This project addresses how E. coli colonizes the large intestine. The data illuminate cellular processes -- nutrition, stress-tolerance, and virulence factors -- that are important for colonization and pathogenesis. Importantly, we found that different E. coli biotypes compete for different sugars in the intestine, suggesting a mechanism for the succession of E. coli strains in healthy individuals and infection by E. coli pathogens that must overcome the colonization resistance barrier imparted by the resident E. coli. In addition, we learned that E. coli must respire oxygen to be competitive in the intestine. This finding suggests that E. coli scavenges oxygen to create anaerobic conditions that favor growth of the anaerobes that dominate the intestinal microbiota. We now have funding to explore and characterize the symbiotic relationship between E. coli and the anaerobes that degrade complex polysaccharides, which in turn release simple sugars to cross-feed E. coli. Our results are explained by the restaurant hypothesis, which predicts microhabitats within the colonic mucus layer where facultative anaerobes, like E. coli, grow on sugars that are served locally by polysaccharide degrading anaerobes.
Single-nucleotide Resolution Transcriptome Analysis. The goal of this project is to understand how bacterial cells work, from genome to transcriptome to metabolome, and to know their genetic circuitry and metabolic networks. To that end, work in my laboratory centers on how colonized bacterial cells grow and compete for nutrients in the microbial community of the mammalian intestine. Two major projects in the laboratory, both focused on E. coli, involve elucidating its symbiotic relation with the anaerobic microbiota in a mouse model of intestinal colonization and generating a single-nucleotide-resolution transcriptome map. Our RNA-Seq transcriptome map builds on the first published E. coli DNA microarray paper and the world’s largest E. coli gene expression database (GenExpDB). In the bioinformatics arena we developed a database schema and middleware, interfaced to intuitive displays, as a computational environment that we have used for discovery of genetic circuits controlling the general stress response and stringent response.
E. coli Gene Expression Database
GenExpDB has a new home at Oklahoma State University! https://genexpdb.okstate.edu
GenExpDB is the world’s largest repository for E. coli gene expression data. This site is a widely used public resource for gene expression analysis.
Current Undergraduate Researchers
 |
Name: Aaron Veenis |
|
 |
Name: Abbie Hoehner |
|
 |
Name: Alisha Beckford |
|
 |
Name: Brooklyn Mossier |
|
 |
Name: Ferris Kalidy |
|
 |
Name: Kassidy Ford (Niblack Scholar) |
|
 |
Name: Matthew Corley |
|
 |
Name: Moses Betru |
|
 |
Name: Nicholas Claassen |
|
 |
Name: Savannah Martin (Wentz Scholar) |
|
 |
Name: Justin Bowen |
Former Undergraduate Researchers
 |
Hannah Sanders | |
 |
Amanda Demackiewicz | |
 |
Amber Eick | |
 |
Kayla Kifer | |
 |
W. Tanner Cole |
Publications (total:116)
- Schmidt, F., Zimmerman, J., Tanna, T., Farouni, R., Conway, T. MacPherson, A. J., Platt, R. J. 2022, Non-invasive assessment of gut function with transcriptional recording sentinel cells. Science 375: 6594: eabm6038. PMID: 35549411
- Conway, T. 2021, Aspartate in the intestine: Dual service as anaerobic electron acceptor and nitrogen source. Environ Microbiol 23(5):2364-65. PMID: 33893749
- King, M., Kubo, A., Kafer, L., Braga, R., Mcleod, D., Khanam, S., Conway, T., and Patrauchan, M. 2021, Calcium regulated protein, CarP, responds to multiple host signals and mediates calcium regulation of Pseudomonas aeruginosa virulence. Appl Environ Microbiol 87(10): e00061-21. PMID: 33674436
- Wu L., Lyu Y., Srinivasagan R., Wu J., Ojo B., Tang M., El-Rassi GD., Metzinger K., Smith BJ., Lucas E.A., Clarke S.L., Chowanadisai W., Shen X., He H., Conway T., von Lintig J., Lin D. 2020, Astaxanthin-Shifted Gut Microbiota Is Associated with Inflammation and Metabolic Homeostasis in Mice. J Nutr 150(10): 2687-2698. PMID: 32810865
- Mokszycki, M. C., Leatham-Jensen, M., Steffensen, J. L., Zhang, Y., Krogfelt, K. A., Caldwell, M. E., Conway, T., and Cohen, P. S. 2018. A Simple In Vitro Gut Model for Studying the Interaction between Escherichia coli and the Intestinal Commensal Microbiota in Cecal Mucus. Appl Environ Microbiol. 84(24): e02166-18. PMID: 30291119
- Kröger, C., MacKenzie, K., Alshabib, E., Kirzinger, M., Suchan, D., Chao, Tzu-C., Akulova, V., Miranda-CasoLuengo, A., Monzon, V., Conway, T., Sivasankaran, S., Hinton, J., Hokamp, K., and Cameron, A. 2018.The primary transcriptome, small RNAs, and regulation of antimicrobial resistance in Acinetobacter baumannii. Nucleic Acids Research. 46(18):9684-9698. PMID: 29986115
- Conway, T. and Cohen, PS. 2015. Applying the restaurant hypothesis to intestinal microbiota. Microbe. 10(8): 324-8.
- Conway, T. and Cohen, PS. 2015. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiol Spectr. 3(3) doi: 10.1128/microbiolspec.MBP-0006-2014. PMID: 26185077
- Schinner S.A., Mokszycki M.E., Adediran J., Leatham Jensen M., Conway T., Cohen P.S. 2015. Escherichia coli EDL933 Requires Gluconeogenic Nutrients to Successfully Colonize the Intestines of Streptomycin-Treated Mice Pre-colonized with E. coli Nissle 1917. Infect Immun. 83:1983-1991. PMID: 25733524
- Creecy, J. P. and T. Conway. 2015. Quantitative bacterial transcriptomics with RNA-seq. Curr Opin Microbiol. 2015, 23:133–140. PMID: 25483350
- Otsuka Y, Muto A, Takeuchi R, Okada C, Ishikawa M, Nakamura K, Yamamoto N, Dose H, Nakahigashi K, Tanishima S, Suharnan S, Nomura W, Nakayashiki T, Aref WG,Bochner BR, Conway T, Gribskov M, Kihara D, Rudd KE, Tohsato Y, Wanner BL, Mori H. 2014. GenoBase: comprehensive resource database of Escherichia coli K-12. Nucleic Acids Res. Nov 15. pii: gku1164 (Epub). PMID: 25399415
- Conway T., Creecy J.P., Maddox S.M., Grissom J.E., Conkle T.L., Shadid T.M., Teramoto J., San Miguel P., Shimada T., Ishihama A., Mori H., Wanner B.L. Unprecedented highresolution view of bacterial operon architecture revealed by RNA sequencing. MBio. 2014 Jul 8;5(4):e01442-14. PMID: 25006232
- Meador JP, Caldwell ME, Cohen PS, Conway, T. 2014. Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect Immun. 82(5): 1931-1938. PMID: 24566621
- Adediran, J., M. Leatham-Jensen, M.E. Mokszycki, J. Frimodt-Møller, K.A. Krogfelt, K. Kazmierczak, L.J. Kenney, T. Conway, and P.S. Cohen. 2014. An Escherichia coli Nissle 1917 envZ Missense Mutant Colonizes the Streptomycin-Treated Mouse Intestine Better than the Wildtype but is not a Better Probiotic. Infect. Immun. 82:670-682. PMID:
24478082 - Kröger, C., A. Colgan, S. Srikumar, K. Händler, S.K. Sivasankaran, D.L. Hammarlöf, R. Canals, J.E. Grissom, T. Conway, K. Hokamp, and J.C. Hinton. 2013. An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14:683-695. PMID: 24331466
- Bertin Y., F. Chaucheyras-Durand, C. Robbe-Masselot, A. Durand, A. de la Foye, J. Harel, P.S. Cohen, T. Conway, E. Forano, C. Martin. 2012. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol 15(2):610-22. PMID: 23126484
- Maltby, R., M.P. Leatham-Jensen, T. Gibson, P.S. Cohen, and T. Conway. 2013. Nutritional Basis for Colonization Resistance by Human Commensal Escherichia coli Strains HS and Nissle 1917 against E. coli O157:H7 in the Mouse Intestine. PLoS One 8(1):e53957. PMID: 23349773.
- Leatham-Jensen, M.P., J. Frimodt-Møller, J. Adediran, M.E. Mokszycki, M.E. Banner, J.E. Caughron, K.A. Krogfelt, T. Conway, P.S. Cohen. 2012. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect Immun 80:1716-27. PMID: 22392928
- Pi, H., S.A. Jones, L.E. Mercer, J.P. Meador, J.E. Caughron, L. Jordan, S.M. Newton, T. Conway, P.E. Klebba. 2012. Role of Catecholate Siderophores in Gram-Negative Bacterial Colonization of the Mouse Gut. PLoS One 7(11):e50020. PMID: 23209633
- Maddox, S.M., P.S. Coburn, N. Shankar, and T. Conway. 2012. Transcriptional Regulator PerA Influences Biofilm-Associated, Platelet Binding, and Metabolic Gene Expression in Enterococcus faecalis. PLoS One 7(4): e34398. PMID: 22496800
- Jones S. A., T. Gibson, R. C. Maltby, F. Chowdhury, V. Stewart, P. S. Cohen, T. Conway. 2011. Anaerobic respiration of Escherichia coli in the mouse intestine. Infect Immun. 79:4218- 26. PMID: 21825069
- Fabich, A. J., M. P. Leatham, J. E. Grissom, G. Wiley, H. Lai, F. Najar, B. A. Roe, P. S. Cohen, and T. Conway. 2011. Genotype and phenotypes of an intestine-adapted Escherichia coli K-12 mutant selected by animal passage for superior colonization. Infect Immun. 79:2430-2439. PMID: 21422176
- Traxler, M. F., M. Zacharia, S. Marquardt, S. M. Summers, H.-T. Nguyen, S. E. Stark and T. Conway. 2011. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 79:830-845. PMID: 21299642.
- Conway, T., and Cohen P.S. 2010. E. coli metabolism in the intestine. Nova Acta Leopoldina 111:103-110.
- Jacobsen, L., L. Durso, T. Conway, and K. W. Nickerson. 2009. Escherichia coli O157:H7 and other E. coli strains share physiological properties associated with intestinal colonization.
Appl Environ Microbiol 75:4633-5. PMID: 19411408 - Leatham, M. P., S. Banerjee, S. M. Autieri, R. Mercado-Lubo, T. Conway, and P. S Cohen. 2009. Pre-colonized Human Commensal E. coli Strains Serve as a Barrier to E. coli
O157:H7 Growth in the Mouse Intestine. Infect Immun 77:2876-86. PMID: 19364832. - Snider, T. A., A. J. Fabich, T. Conway, and K. D. Clinkenbeard. 2008. E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet Microbiol.
36:150-4. PMID: 19095384 - Mercado-Lubo R., M. P. Leatham, T. Conway, P. S. Cohen. 2009. Salmonella enterica serovar Typhimurium mutants unable to convert malate to pyruvate and oxaloacetate are
avirulent and immunogenic in BALB/c mice. Infect Immun 77:1397-405. PMID: 19168732 - Hu J.C., R. Aramayo, D. Bolser, T. Conway, C.G. Elsik, M. Gribskov, T. Kelder, D. Kihara, T.F. Knight Jr, A.R. Pico, D.A. Siegele, B. L. Wanner, R. D. Welch. 2008. The emerging
world of wikis. Science. 320(5881):1289-90. PMID: 18535227 - Jones, S. A., M. Jorgensen, F. Z. Chowdhury, R. Rodgers, J. Hartline, M. P. Leatham, C. Struve, K. A. Krogfelt, P. S. Cohen, and T. Conway. 2008. Glycogen and Maltose Utilization by
Escherichia coli O157:H7 in the Mouse Intestine. Infect Immun 76:2531-40. PMID: 18347038 - Traxler, M. F., S. M. Summers, H. T. Nguyen, V. M. Zacharia, G. A. Hightower, J. T. Smith, and T. Conway. 2008. The global, ppGpp-mediated stringent response to amino acid
starvation in Escherichia coli. Mol Microbiol 68:1128-48. PMID: 18430135. Citations: 83 - Mercado-Lubo, R., E. J. Gauger, M. P. Leatham, T. Conway, and P. S. Cohen. 2008. A Salmonella enterica serovar Typhimurium Succinate Dehydrogenase/Fumarate
Reductase Double Mutant is Avirulent and Immunogenic in BALB/c Mice. Infect Immun 76:1128-34. PMID: 18086808 - Fabich, A. J., S. A. Jones, F. Z. Chowdhury, A. Cernosek, A. Anderson, D. Smalley, J. W. McHargue, G. A. Hightower, J. T. Smith, S. M. Autieri, M. P. Leatham, J. J. Lins, R. L.
Allen, D. C. Laux, P. S. Cohen, and T. Conway. 2008. Comparative carbon nutrition of pathogenic and commensal Escherichia coli in the mouse intestine Infect Immun
76:1143-52. (Selected for “Editor’s Spotlight”) PMID: 18180286 - Autieri, S. M., J. J. Lins, M. P. Leatham, D. C. Laux, T. Conway, and P. S. Cohen. 2007. LFucose Stimulates Utilization of D-Ribose by Escherichia coli MG1655 Delta-fucAO and
E. coli Nissle 1917 Delta-fucAO Mutants in the Mouse Intestine and in M9 Minimal Medium. Infect Immun. 75:5465-75. (Selected for “Editor’s Spotlight”) PMID: 17709419 - Jones, S. A., F. Z. Chowdhury, A. J. Fabich, A. Anderson, D. M. Schreiner, A. L. House, S. M. Autieri, M. P. Leatham, J. J. Lins, M. Jorgensen, P. S. Cohen, and T. Conway. 2007.
Respiration of Escherichia coli in the Mouse Intestine. Infect Immun 75:4891-9. (Selected for “Editor’s Spotlight”) PMID: 17698572 - Gauger, E. J., M. P. Leatham, R. Mercado-Lubo, D. C. Laux, T. Conway, P. S. Cohen. 2007. Role of Motility and the flhDC Operon in Escherichia coli MG1655 Colonization of the
Mouse Intestine. Infect Immun. 75:3315-24. PMID: 17438023 - Snider, T. A., A. J. Fabich, K. E. Washburn, W. P. Sims, J. L. Blair, P. S. Cohen, T. Conway, and K. D. Clinkenbeard. 2006. Evaluation of a model for Escherichia coli O157:H7
colonization in streptomycin-treated adult cattle. Am J Vet Res. 67:1914-20. PMID: 17078755 - Wren J.D. and T. Conway. 2006. Meta-Analysis of Published Transcriptional and Translational Fold-changes Reveals a Bias Towards Low-fold Induction. OMICS. 10(1):15-27.
PMID: 16584315 - Traxler, M. F., D.-E. Chang, and T. Conway. 2006. Guanosine 5’,3’-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli.
Proc. Nat. Acad. Sci. 103: 2374-2379. PMID: 16467149 - Tchawa Yimga M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella
enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74:1130-40 PMID: 16428761 - Wren J.D., J.E. Grissom, and T. Conway. 2006. Email decay rates among MEDLINE corresponding authors. EMBO Reports. 7: 122-127. PMID: 16452921
- Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. The mouse intestine selects stable nonmotile
flhDC operon deletion mutants of Escherichia coli MG1655 with increased mouse large intestine colonizing ability and more efficient utilization of carbon sources. Infect.
Immun. 73: 8039-8049. PMID: 16299298 - Murray, E. L. and T. Conway. 2005. Multiple regulators control expression of the Entner- Doudoroff aldolase (Eda) of Escherichia coli. J. Bacteriol. 187: 991-1000. PMID:
15659677 - Santosa, B., T. Conway, and T.B. Trafalis. 2005. A hybrid knowledge based-clustering multiclass SVM approach for genes expression analysis. Biomedicine 2005
Bates Utz, C., A. B. Nguyen, D. J. Smalley, A. B. Anderson, and T. Conway. 2004. GntP is the Escherichia coli Fructuronic Acid Transporter and Belongs to the UxuR Regulon. J.
Bacteriol. 186:7690-7696. PMID: 15516583 - Wren, J. D., M. Yao, M. Langer, and Tyrrell Conway. 2004. Simulated annealing of microarray data reduces noise and enables cross-experimental comparisons. DNA Cell Biol. 23: 695-
700. PMID: 15585127 - Chang, D.E., D. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. Carbon nutrition of
Escherichia coli in the mouse intestine. 2004. Proc. Nat. Acad. Sci. 101: 7427–7432. PMID: 15123798 - Miranda, R. L., T. Conway, M. P. Leatham, D. E. Chang, D. L. Tucker, W. E. Norris, J. H. Allen, S. J. Stevenson, D. C. Laux, and P. S. Cohen. 2004. Glycolytic and
gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 72: 1666-76. PMID: 14977974 - Bausch, C., M. Ramsey, and T. Conway. 2004. Transcriptional Organization and Regulation of the L-Idonic Acid Pathway (GntII System) In Escherichia coli. J. Bacteriol. 186: 1388-
97. PMID: 14973046 - Ma , Z., S. Gong, D. L.Tucker, T. Conway, and J. W Foster. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K12. Mol.
Microbiol. 49: 1309-1320. PMID: 12940989 - Wanken, A., T. Conway, and K. A. Eaton. 2003. The Entner-Doudoroff pathway has little effect on Helicobacter pylori colonization of mice. Infect. Immun. 71: 2920-2923.
PMID: 12704170 - Wolfe, A. J., D.E. Chang, J. D. Walker, J. E. Seitz-Partridge, M. D. Vidaurri, B. Pruess, M. C. Henk, J. C. Larkin, and T. Conway. 2003. Evidence that acetyl phosphate functions as a
global signal during biofilm development. Mol. Microbiol. 48: 977-988. PMID: 12753190 - Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in E. coli. J. Bacteriol. 185: 3190-3201.
PMID: 12730179 - Wei, N., L. Gruenwald, and T. Conway. 2003. Analyzing Escherichia coli gene expression data by a multilayer adjusted tree organizing map. The 3rd IEEE Symposium on
Bioinformatics and Bioengineering, Bethesda MD, March 10-12, 2003, pp. 289-296. - Møller, A., T. Conway, P. Nuijten, K. A. Krogfelt and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal
mucus, but fails to colonize the mouse large intestine. Infect. Immun. 71: 2142-2152. PMID: 12654836 - Conway, T. and G. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47: 879-889. (Top 50 download at Mol
Micro) PMID: 12581346 - Smalley, D. J., M. Whiteley, and T. Conway. 2003. In search of the minimal E. coli genome. Trends Microbiol. 11: 6-8. PMID: 12526847
- Ma, Z., H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2002. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators,
GadX and GadW. J. Bacteriol. 184: 7001-7012. PMID: 12446650 - Tucker, D. L., N. Tucker, T. Conway. 2002. Gene expression profiling of the pH response in E. coli. J. Bacteriol. 183: 6551-6558. PMID: 12426343
- Santosa, B., T. Conway, and T. B. Trafalis. 2002. Knowledge based-clustering and application of multi-class SVM for gene expression analysis. Conference paper: ANNIE2002, Smart
Engineering System Design, November 10-13, 2002, St. Louis, MO; Publication: IN Intelligent Engineering Systems Through Artificial Neural Networks, Dagli, C. H., A. L. - Buczak, J. Ghosh, M. J. Embrchts, O. Ersoy, S. W. Dercel (eds.), ASME Press 12: 391-395.
- Chang, D.E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of E. coli growth transitions: an expanded stringent response model. Mol Microbiol 45: 289-306. PMID:
12123445 - Conway, T., B. Kraus, D. L. Tucker, D. J. Smalley, A. F. Dorman, and L. McKibben. 2002. DNA array analysis in a microsoft windows environment. Biotechniques. 32: 110-119.
PMID: 11808684 - Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol.
181: 6425-6440. PMID: 10515934 - Matsuno, K., T. Blais, A. W. Serio, T. Conway, T. M. Henkin, and A. L. Sonenshein. 1999. Metabolic imbalance and sporulation in an isocitrate dehydrogenase mutant of Bacillus
subtilis. J. Bacteriol. 181: 3382-3291. PMID: 10348849 - Peekhaus, N. and T. Conway. 1998. What’s for Dinner? Entner-Doudoroff metabolism in Escherichia coli. (A mini-review) J. Bacteriol. 180: 3495-3502. PMID: 9657988
- Bausch, C., N. Peekhaus, C. Utz, T. Blais, E. Murray, T. Lowary, and T. Conway. 1998. Sequence Analysis of the GntII (Subsidiary) System for Gluconate Metabolism Reveals a
Novel Pathway for L-Idonic Acid Catabolism in E. coli. J. Bacteriol. 180: 3704-3710. PMID: 9658018 - Peekhaus, N. and T. Conway. 1998. Positive and negative transcriptional regulation of the gntT gene of the gluconate regulon of Escherichia coli by GntR and the CRP-cAMP complex.
J. Bacteriol. 180: 1777-1785. PMID: 9537375 - Fuhrman, L. K., A. Wanken, K. W. Nickerson, and T. Conway. 1998. Rapid accumulation of intracellular 2-keto-3-deoxy-6-phosphogluconate in an Entner-Doudoroff aldolase mutant
results in bacteriostasis. FEMS Microbiol. Letts. 159: 261-266. PMID: 9503620 - Parker, C., N. Peekhaus, X. Zhang, and T. Conway. 1997. Kinetics of sugar transport and phosphorylation influence glucose and fructose cometabolism by Zymomonas mobilis.
Appl. Environ. Microbiol. 63: 3519-3525. PMID: 16535690 - Porco, A., N. Peekhaus, C. Bausch, S. Tong, T. Isturiz, and T. Conway. 1997. Molecular genetic characterization of the Escherichia coli gntT gene of GntI, the main system for
gluconate metabolism. J. Bacteriol. 179: 1584-1590. PMID: 9045817 - Peekhaus, N., S. Tong, E. Murray, J. R. Reizer, M. Saier, and T. Conway. 1997. Characterization of a novel transporter family that includes multiple Escherichia coli
gluconate transporters and their homologs. FEMS Microbiol. Letters 147: 233-238. PMID: 9119199 - Wahlund, T. M., T. Conway, and F. R. Tabita. 1996. Bioconversion of CO2 to ethanol and other compounds. Amer. Chem Soc. Div. Fuel Chem. 41 (4): 1403-1406.
Gold, R. S., M. M. Meagher, R. W. Hutkins, S. Tong, and T. Conway. 1996. Cloning and expression of the Zymomonas mobilis "production of ethanol" genes in Lactobacillus
casei. Curr Microbiol. 33: 256-260. PMID: 8824172 - Snoep, J. L., N. Arfmann, L. P. Yomano, H. V. Westerhoff, T. Conway, and L. O. Ingram. 1996. Control of glycolytic flux in Zymomonas mobilis by gene products from the glf-zwf-eddglk
operon. Biotechnol. Bioeng. 51: 190-197. PMID: 18624328 - Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147: 448-455. PMID: 9084754
- Tong, S., A. Porco, T. Isturiz, and T. Conway. 1996. Cloning and molecular characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate
metabolism. J. Bacteriol. 178: 3620-3269. PMID: 8655507 - Klemm, P., S. Tong, H. Nielsen, and T. Conway. 1996. The gntP gene of Escherichia coli involved in gluconate uptake. J. Bacteriol. 178:61-67. PMID: 8550444
- Bhattacharya, M., L. Fuhrman, A. Ingram, K. W. Nickerson, and T. Conway. 1995. Single-run separation and detection of multiple metabolic intermediates by anion exchange-HPLC
and application to cell pool extracts prepared from Escherichia coli. Analytical Biochemistry 232: 98-106. PMID: 8600840 - Parker, C. T., W. O. Barnell, J. L. Snoep, L. O. Ingram, and T. Conway. 1995. Characterization of the Zymomonas mobilis glucose facilitator gene product (glf) in recombinant
Escherichia coli: Examination of transport mechanism, kinetics, and the role of glucokinase in glucose transport. Molecular Microbiol. 15: 795-802. PMID: 7596282 - Snoep, J. L., N. Arfman, L. P. Yomano, R. K. Fliege, T. Conway, and L. O. Ingram. 1994. Reconstitution of glucose uptake and phosphorylation in a glucose negative mutant of
Escherichia coli by using the Zymomonas mobilis genes encoding the glucose facilitator protein and glucokinase. J. Bacteriol. 176: 2133-2135. PMID: 8144485 - Fliege, R., S. Tong, A. Shibata, K. W. Nickerson, and T. Conway. 1992. The Entner-Doudoroff pathway in Escherichia coli is induced for oxidative glucose metabolism via
pyrroloquinoline quinone-dependent glucose dehydrogenase. Appl. Environ. Microbiol. 58: 3826-3829. PMID: 1335716 - Mejia, J.P., M.E. Burnett, H. An, W.O. Barnell, K.F. Keshav, T. Conway and L.O. Ingram. 1992. Coordination of Zymomonas mobilis glycolytic and fermentative enzymes: A
simple hypothesis based on mRNA stability. J. Bacteriol. 174: 6438-6443. PMID: 1400196 - Burnett, M.E., J. Liu, and T. Conway. 1992. Molecular characterization of the Zymomonas mobilis enolase (eno) gene. J. Bacteriol. 174: 6548-6553. PMID: 1400207
- Shark, K. and T. Conway. 1992. Cloning and molecular characterization of the DNA ligase gene (lig) from Zymomonas mobilis. FEMS Microbiol. Letts. 96: 19-26. PMID: 1526462
- Egan, S., R. Fliege, S. Tong, A. Shibata, R.E. Wolf, Jr. and T. Conway. 1992. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: Sequence analysis
and localization of promoters for the edd-eda operon. J. Bacteriol. 174: 4638-4646. PMID: 1624451 - Zembrzuski, B., P. Chilco, S.-L. Liu, J. Liu, T. Conway, and R. K. Scopes. 1992. The fructokinase gene from Zymomonas mobilis: Cloning, sequencing, expression and
structural comparison with other hexose kinases. J. Bacteriol. 174: 3455-3460. PMID: 1317376 - Gold, S., M. M. Meagher. R. Hutkins, and T. Conway. 1992. Ethanol tolerance and carbohydrate metabolism in Lactobacilli. Journal for Industrial Microbiology 10:45-54.
- Barnell, W. O., J. Liu, T. L. Hesman, M. C. O'Neill, and T. Conway. 1992. The Zymomonas mobilis glf, zwf, edd, and glk genes form an operon: Localization of the promoter and
identification of a conserved sequence in the regulatory region. J. Bacteriol. 174: 2816-2823. PMID: 1569013 - Liu, J., W. O. Barnell, and T. Conway. 1992. The polycistronic mRNA of the Zymomonas mobilis glf-zwf-edd-glk operon is subject to complex transcript processing. J. Bacteriol.
174: 2824-2833. PMID: 1569014 - Conway, T., R. Fliege, D. Jones-Kilpatrick, J. Liu, W.O. Barnell, and S. E. Egan. 1991. Cloning, characterization, and expression of the Zymomonas mobilis eda gene that
encodes 2- keto-3-deoxy-6-phosphogluconate aldolase of the Entner-Doudoroff pathway. Molec. Microbiol. 5: 2901-2911. PMID: 1809834 - Conway, T., K.C. Yi, S.E. Egan, R.E. Wolf, and D.L. Rowley. 1991. Location of the zwf, edd, and eda genes on the Escherichia coli physical map. J. Bacteriol. 173: 5247-5248.
PMID: 1885506 - Adamowicz, M., T. Conway and K. Nickerson. 1991. Nutritional restoration of oxidative glucose metabolism in Escherichia coli by addition of the glucose dehydrogenase
cofactor PQQ. Appl. Environ. Microbiol. 57: 2012-2015. PMID: 1654044 - Hesman, T.L., W.O. Barnell and T. Conway. 1991. Cloning, characterization and sequence analysis of a phosphoglucose isomerase gene (pgi) from Zymomonas mobilis that is
subject to carbon source-dependent regulation. J. Bacteriol. 173: 3215-3223. PMID: 1708765 - Lal, S.K., S. Johnson, T. Conway and P.M. Kelley. 1991. Characterization of a Maize cDNA which complements an enolase deficient mutant of Escherichia coli. Plant Molec. Biol.
16: 787-795. PMID: 1859865 - Barnell, W.O., K.C. Yi, and T. Conway. 1990. Sequence and genetic organization of a Zymomonas mobilis gene cluster that encodes several enzymes of glucose metabolism. J.
Bacteriol. 172: 7227-7240. PMID: 2254282 - Eddy, C.K., J.P. Mejia, T. Conway, and L.O. Ingram. 1989. Differential expression of gap and pgk genes within the gap operon of Zymomonas mobilis. J. Bacteriol. 171: 6549-6554.
PMID: 2687242 - MacKenzie, K.F., T. Conway, H.C. Aldrich, and L.O. Ingram. 1989. Expression of alcohol dehydrogenase II and adhB-lacZ operon fusions in recombinant Zymomonas mobilis. J.
Bacteriol. 171: 4577-4582. PMID: 2504692 - Conway, T. and L.O. Ingram. 1989. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and
Saccharomyces cerevisiae. J. Bacteriol. 171: 3754-3759. PMID: 2661535 - Pond, J.L., C.K. Eddy, K.F. MacKenzie, T. Conway, D.J.Borecky, and L.O. Ingram. 1989. Cloning, sequencing and characterization of the principal acid phosphatase, the phoC+
product, from Zymomonas mobilis. J. Bacteriol. 171: 767-774. PMID: 2914872 - Conway, T. and L.O. Ingram. 1988. Phosphoglycerate kinase from Zymomonas mobilis: Cloning, sequencing and localization to the gap operon. J. Bacteriol. 170: 1926-1933.
PMID: 2832389 - Ingram, L.O. and T. Conway. 1988. Expression of different levels of ethanologenic enzymes from Zymomonas mobilis in recombinant strains of E. coli. Appl. Environ. Microbiol.
54: 397- 404. PMID: 16347553 - Conway, T., G.W. Sewell, and L.O. Ingram. 1987. Glyceraldehyde phosphate dehydrogenase from Zymomonas mobilis: Cloning, sequencing and identification of promoter region. J.
Bacteriol. 169: 5653-5662. PMID: 3680173 - Ingram, L.O., T. Conway, D.P. Clark, G.W. Sewell, and J.F. Preston. 1987. Genetic engineering of ethanol production in E. coli. Appl. Environ. Microbiol. 53: 2420-2425. PMID:
3322191 - Osman, Y.A., T. Conway, S.J. Bonetti, and L.O. Ingram. 1987. Glycolytic flux in Zymomonas mobilis: Enzyme and metabolite levels during batch fermentation. J. Bacteriol. 169:
3726- 3736. PMID: 3611027 - Conway, T., G.W. Sewell, Y.A. Osman, and L.O. Ingram. 1987. Cloning and sequencing of alcohol dehydrogenase II from Zymomonas mobilis. J. Bacteriol. 169: 2591-2597.
PMID: 3584063 - Conway, T., Y.A. Osman, and L.O. Ingram. 1987. Gene expression in Zymomonas mobilis: Promoter structure and identification of membrane anchor sequences forming functional
LacZ' fusion proteins. J. Bacteriol. 169: 2327-2335. PMID: 3034853 - Conway, T., Y.A. Osman, J.I. Konnan, E.M. Hoffmann, and L.O. Ingram. 1987. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J. Bacteriol.
169: 949-954. PMID: 3029037 - Conway, T., M. O-K. Byun, and L.O. Ingram. 1987. Expression vector for Zymomonas mobilis. Appl. Environ. Microbiol. 53: 235-241. PMID: 16347272
- Conway T, Cohen PS. 2014. Commensal and pathogenic Escherichia coli metabolism in the gut. In Conway T, Cohen PS (ed.), Metabolism and Bacterial Pathogenesis, IN PRESS. ASM Press, Washington, DC.
- Conway, T., and Cohen P.S. 2007. Escherichia coli at the Intestinal Mucosal Surface. In: 4th Edition Virulence Mechanisms of Bacterial Pathogens.
- K.A. Brogden, F.C. Minion, N. Cornick, T.B. Stanton, Q. Zhang, L.K. Nolan, and M.J. Wannemuehler (eds.), American Society for Microbiology Press, Washington, D. C., pp. 175-196, 2007.
- Han, J., L. Gruenwald, and T. Conway. 2005. Data mining in gene expression data analysis: a survey. In: Managing Complex Data for Decision Support, Darmont, J. and O. Boussaid,
eds., Idea Group, Inc., Hershey, PA. - Chang, D.E. and T. Conway. 2005. Metabolic genomics. 50: 1-39. In: Advances in Microbial Physiology, R. K. Poole, ed., Elsevier, Amsterdam. PMID: 16221577
- Laux, D. C., P. S. Cohen, and T. Conway. 2005. Role of the mucus layer in bacterial colonization of the intestine. In: Colonization of Mucosal Surfaces, Nataro, J. P., H. L. T.
Mobley, and P. S. Cohen, eds., ASM Press, Washington, DC. - Conway, T., K. A. Krogfelt, and P. S. Cohen. 29 December 2004, posting date. Chapter 8.3.1.2, The life of commensal Escherichia coli in the mammalian intestine. In: R. Curtis III,
editor in chief, EcoSal – Escherichia coli and Salmonella: Cellular and Molecular Biology [3rd edition, Online.] http://www.ecosal.org. ASM Press, Washington, D.C. - Conway, T. 1992. The Entner-Doudoroff pathway: History, physiology, and molecular biology. FEMS Microbiol. Rev. 103: 1-27. PMID: 1389313
- Ingram, L.O., C.K. Eddy, K.F. MacKenzie, T. Conway, and F. Alterthum. 1989. Genetics of Zymomonas mobilis and ethanol production. Develop. Indust. Microbiol. 30: 53-69.
- Ingram, L.O., Y.A. Osman, and T. Conway. 1988. Zymomonas mobilis: Effects of ethanol on fermentation. In, N. van Uden (ed.), Alcohol Toxicity in Yeast and Bacteria. CRC Press.
U.S. Patents Issued
Conway, T., L.O. Ingram, and F. Alterthum. 1991. Ethanol Production by Escherichia coli
Strains Co-expressing Zymomonas PDC and ADH Genes. U. S. Patent No. 5,000,000.
Filed May 15, 1989. Issued March 19, 1991.
Ingram, L. O. and T. Conway. 1992. Cloning and Sequencing of the Alcohol Dehydrogenase II
Gene from Zymomonas mobilis. U.S. Patent No. 5,162,516. Filed May 31, 1988. Issued
November 10, 1992.